This is another recent paper using a large number of genetic markers to study genetic structure within a race. You can read the paper for yourselves, but some comments are in order.
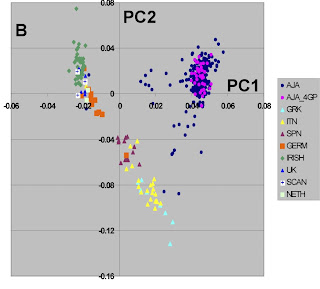
First, this mirrors the results of another recent study on European-American population structure. Mediterranean populations are distinguished from Northern European populations and from Jews.
Doubtlessly, the discontinuities between the three groups evident in the Figure would be less clear if more populations were sampled, but nonetheless this is clear evidence that not only race, but ethnicity too has a genetic component.
If e.g., we were given the DNA of a German, a Greek, and a Jew, we would be able to tell them apart with very high probability. All it would take, would be to genotype them for the 300K SNPs, and subsequently score them for the first two principal components.
Another interesting feature of this study is the form of the Jewish cluster. The purple dots correspond to Jewsh with all four Jewish grandparents and they form a tight cluster. This is not the case when less stringent criteria (blue dots) are used.
This is a wonderful visual display of what happens to a partially inbred group (Ashkenazi Jews) as a result of intermixture. As long as within-group marriage patterns prevail, genetic similarity is maintained, but intermarriage leads to an attenuation of the group's genetic distinctiveness.
It is interesting to think of how this picture will change as genetic science moves forward:
If more populations are sampled, then the gaps between populations may "fill up". I would not be surprised, for example, if the French would occupy the gap between Spaniards and Germans.
If more SNPs were used, then populations that overlap greatly (say Greeks and Italians) might be separated.
Ethnic groups may differ both in culture and in genes, but these two dimensions of ethnicity are not independent. Differences in culture are barriers to gene flow; over time, such barriers differentiate gene pools. Conversely, genetic intermixture bring separate gene pools closer together, but also serves to homogenize cultures.
PLoS Genetics Vol. 4, No. 1, e4 doi:10.1371/journal.pgen.0040004
Analysis and Application of European Genetic Substructure Using 300 K SNP Information
Tian C, Plenge RM, Ransom M, Lee A, Villoslada P, et al.
European population genetic substructure was examined in a diverse set of >1,000 individuals of European descent, each genotyped with >300 K SNPs. Both STRUCTURE and principal component analyses (PCA) showed the largest division/principal component (PC) differentiated northern from southern European ancestry. A second PC further separated Italian, Spanish, and Greek individuals from those of Ashkenazi Jewish ancestry as well as distinguishing among northern European populations. In separate analyses of northern European participants other substructure relationships were discerned showing a west to east gradient. Application of this substructure information was critical in examining a real dataset in whole genome association (WGA) analyses for rheumatoid arthritis in European Americans to reduce false positive signals. In addition, two sets of European substructure ancestry informative markers (ESAIMs) were identified that provide substantial substructure information. The results provide further insight into European population genetic substructure and show that this information can be used for improving error rates in association testing of candidate genes and in replication studies of WGA scans.
Link

No comments:
Post a Comment