August 31, 2009
Eye-tracking of men's preferences for women's body shape
Eye-Tracking of Men's Preferences for Waist-to-Hip Ratio and Breast Size of Women.
Dixson BJ, Grimshaw GM, Linklater WL, Dixson AF.
Studies of human physical traits and mate preferences often use questionnaires asking participants to rate the attractiveness of images. Female waist-to-hip ratio (WHR), breast size, and facial appearance have all been implicated in assessments by men of female attractiveness. However, very little is known about how men make fine-grained visual assessments of such images. We used eye-tracking techniques to measure the numbers of visual fixations, dwell times, and initial fixations made by men who viewed front-posed photographs of the same woman, computer-morphed so as to differ in her WHR (0.7 or 0.9) and breast size (small, medium, or large). Men also rated these images for attractiveness. Results showed that the initial visual fixation (occurring within 200 ms from the start of each 5 s test) involved either the breasts or the waist. Both these body areas received more first fixations than the face or the lower body (pubic area and legs). Men looked more often and for longer at the breasts, irrespective of the WHR of the images. However, men rated images with an hourglass shape and a slim waist (0.7 WHR) as most attractive, irrespective of breast size. These results provide quantitative data on eye movements that occur during male judgments of the attractiveness of female images, and indicate that assessments of the female hourglass figure probably occur very rapidly.
Link
August 30, 2009
mtDNA and ethnic differentiation in East Africa
The pattern observed in East Africa (with the exception of the Khoisan-related Hadza and Sandawe populations), which combines a high level of within-population diversity with strong genetic structure among populations, suggests the occurrence of periodical episodes of admixture in these populations, separated by periods of isolation and genetic drift. Indeed, the observation of high levels of diversity within populations could be due to long-term large effective population sizes maintained in East Africa. In this case, however, little genetic structure between populations should be expected, since there would be little opportunity for genetic drift to act. Alternatively, gene flow can produce high within population diversity, and in the present case, it could also account for the extensive sharing of haplotypes and haplogroups observed between the Nyangatom and the Daasanach, as well as with other populations.
The intermediate linkage disequilibrium (LD) found in East Africa (Tishkoff et al., 1996) in contrast with Europe (high LD) and Sub-Saharan Africa (low LD, Tishkoff & Kidd, 2004; Conrad et al., 2006), could be due to such admixture events, more frequently occurring in this region compared to other Sub-Saharan populations. Substantial levels of gene flow among Nilo-Saharan, Afro-Asiatic and Niger-Congo populations from Tanzania have already been inferred by Tishkoff et al. (2007a) and our results suggest that these gene flows could have occurred in a larger region extending up to Southern Ethiopia.
The high diversity in East Africa was interpreted as a sign of an ancient origin. However, our results might indicate that this high diversity could also come from a particular history of recent migrations and admixture promoted by the pastoralist societies that dominate in the region.
- They are the oldest population, and have been accumulating genetic diversity for the longest period of time
- They are substantially admixed with very divergent components (e.g., Semites, Nilo-Saharans, Cushitic speakers, and so on)
Genetic Evidence for Complexity in Ethnic Differentiation and History in East Africa.
Poloni ES, Naciri Y, Bucho R, Niba R, Kervaire B, Excoffier L, Langaney A, Sanchez-Mazas A.
Summary
Link
August 28, 2009
Lactase persistence spread with Neolithic Linearbandkeramik
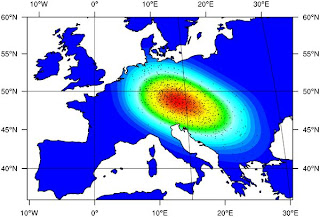 From the paper:
From the paper:Following acceptance at the 0.5% level and regression adjustment we found that the most probable location where an LP allele first underwent selection among dairying farmers lies in a region between the central Balkans and central Europe (see Figure 3). It should be noted that, as simulated, we did not attempt to identify the location where the LP −13,910*T allele first arose. Instead we assumed that it started to rise to appreciable frequencies only after selection began among dairying farmers, initially at the particular location we estimated. The timing of the start of this gene-culture coevolution process was therefore strongly influenced by the arrival time of dairying farmers at the location where selection began in simulations. Since we selected simulations that give a good fit to the timing of the arrival of farming at different locations [31], we estimated a narrow range of dates for when selection began (95% CI 6,256 to 8,683 years BP;In other words, selection for the lactase persistence allele did not result in modern Europeans having a larger proportion of their ancestry from the place where this process began.
...
Although not strictly a parameter of the model presented we have applied the ABC approach to estimate the genetic contribution of people living in the deme where LP-dairying gene-culture coevolution began, and its 8 surrounding demes, to the modern European gene-pool (95% CI 2.83 to 27.4%; mode = 7.47%; see Figure 4B) ... We then compared the distributions of genetic contribution (of people living in and around the LP-dairying start deme to the modern European genepool) with and without selection acting. To our surprise the two distributions are nearly identical.
Perhaps the most interesting result presented here is our estimation of the geographic and temporal origins of LP-dairying co-evolution. We find the highest posterior probabilities for a region between the central Balkans and central Europe (see Figure 3). At first sight such a location of origin may seem counter intuitive since it is far-removed from Northwest Europe, where the −13,910*T allele is found at highest frequency. However, previous simulations have shown that the geographic centroid of allele can be offset from its location of origin, particularly when it occurs on the wave front of a demographic expansion [29],[30]. The lactase-dairying coevolution origin region inferred here is consistent with a number of archaeologically attested patterns concerning the emergence and spread of dairying. Recent carbon isotope ratios from lipids extracted from archaeological sherds show the presence of milk fats in present-day western Turkey and connect these findings to an increased importance of cattle herding [26], [45]–[48]. In general, the spread of the Neolithic lifestyle from the Aegean to Central Europe goes hand in hand with the decline of the importance of sheep and goat and the rise in frequency of cattle bones in archaeological assemblages. While the Balkans at the beginning of the Neolithic still shows a variety of subsistence strategies [49], the middle Neolithic in SE-Europe and the earliest Neolithic in Central Europe after 7,500 BP show a clear preponderance of cattle.
There's only one little problem: It's hard to see how the same scenario gets the allele to India. Or, for that matter, Ireland. The authors posit that Indian lactase persistence will be found to be caused by a "diversity" of alleles. They seem to have missed this paper that found a greater diversity of lactase-associated haplotypes "north of the Caucasus" -- consistent with an initial steppe dispersal. OK, that's two problems, and they're not little.
- Higher allele frequency in northwestern Europe compared to India suggests that the "point of origin" ought to be closer to the former than to the latter, or that the allele's selection began earlier in the former than in the latter.
- We must account for terrain and mode of transmission. The steppelands stretching from eastern Europe to the outskirts of China, combined with the invention of full pastoral nomadism made it possible for the spread of genes at a speed impossible for regular "demic diffusion". Moreover, a great part of this territory was essentially devoid of previous populations, and, the economy of the nomads necessitated its continued positive selection. Thus, the allele's frequency would not have been diluted by the time it reached the eastern ends of its expansion.
- The beginning of selection inferred in the current study is much older than the invention of pastoral nomadism. Inclusion of more populations could only push the time further into the past; it could not make it more recent. Thus, advocates of an "eastern" solution must explain how an allele appears to have started experiencing selection in the geographical region examined in the current paper thousands of years before it was introduced from the east.
- An eastern-western mode of transmission would result in an eastern-western cline, not a northern-southern one. An additional mechanism would need to be invoked to explain the latter.
The Origins of Lactase Persistence in Europe
Yuval Itan et al.
Abstract
Lactase persistence (LP) is common among people of European ancestry, but with the exception of some African, Middle Eastern and southern Asian groups, is rare or absent elsewhere in the world. Lactase gene haplotype conservation around a polymorphism strongly associated with LP in Europeans (−13,910 C/T) indicates that the derived allele is recent in origin and has been subject to strong positive selection. Furthermore, ancient DNA work has shown that the −13,910*T (derived) allele was very rare or absent in early Neolithic central Europeans. It is unlikely that LP would provide a selective advantage without a supply of fresh milk, and this has lead to a gene-culture coevolutionary model where lactase persistence is only favoured in cultures practicing dairying, and dairying is more favoured in lactase persistent populations. We have developed a flexible demic computer simulation model to explore the spread of lactase persistence, dairying, other subsistence practices and unlinked genetic markers in Europe and western Asia's geographic space. Using data on −13,910*T allele frequency and farming arrival dates across Europe, and approximate Bayesian computation to estimate parameters of interest, we infer that the −13,910*T allele first underwent selection among dairying farmers around 7,500 years ago in a region between the central Balkans and central Europe, possibly in association with the dissemination of the Neolithic Linearbandkeramik culture over Central Europe. Furthermore, our results suggest that natural selection favouring a lactase persistence allele was not higher in northern latitudes through an increased requirement for dietary vitamin D. Our results provide a coherent and spatially explicit picture of the coevolution of lactase persistence and dairying in Europe.
Link
Refinement of ancestry informative markers in Europeans (Tian et al. 2009)
In general, Fst values corresponded to geographical relationships with smaller values between population groups with origins in neighboring countries/regions (e.g. Tuscan/Greek, Fst = 0.001) compared with those from very different regions in Europe (e.g. Russian/Palestinian, Fst = 0.020) similar to previous studies [10].The paired Fst table confirms that the closest population to Greeks are Italians (negative Fst=-0.0001) and Tuscans (Fst=0.0005). Much further apart are Spaniards (Fst=0.0035) and Germans (Fst=0.0039), who are still much closer than the most distant Russians (Fst=0.0108) and Orcadians (Fst=0.103).
...
The current study extends the analysis of European population genetic structure to include additional southern European groups and Arab populations. Even within Italy, the relative position of northern Italians compared with subjects from Tuscany is consistent with the general geographic correspondence of PCA results. Interestingly, the majority of Italian Americans (NYCP 4 grandparent defined) appear to derive from southern Italy and overlap with subjects of Greek heritage. Both of these observations are consistent with previous historical information [30,31].
 Some observations:
Some observations:- In Α we see a west-east differentiation in northern Europe, with Irish and Russians in the two ends of PC1.
- In Β we see differentiation of non-Jewish southern European populations from Ashkenazi Jews along PC1 and from Druze, Palestinians, and Bedouins, along PC2. Greeks are concentrated near the center at the lower left quadrant.
- In C we see all the populations using only ancestry-informative markers and in D with all 270k markers. The two plots are similar, although use of the full set results in clearer results. We observe a cline of populations from the Near East to Northern Europe at the bottom. A little discontinuity between Greeks and Arabs would probably disappear if geographically intermediate populations had been included. Ashkenazi Jews are differentiated from the entire sample, suggesting that due to genetic drift, selection, or cryptic other ancestry (?) they cannot be reckoned as a simple European-Near Eastern mix genetically.

European Population Genetic Substructure: Further Definition of Ancestry Informative Markers for Distinguishing Among Diverse European Ethnic Groups.
Tian C, Kosoy R, Nassir R, Lee A, Villoslada P, Klareskog L, Hammarström L, Garchon HJ, Pulver AE, Ransom M, Gregersen PK, Seldin MF.
The definition of European population genetic substructure and its application to understanding complex phenotypes is becoming increasingly important. In the current study using over 4000 subjects genotyped for 300 thousand SNPs we provide further insight into relationships among European population groups and identify sets of SNP ancestry informative markers (AIMs) for application in genetic studies. In general, the graphical description of these principal components analyses (PCA) of diverse European subjects showed a strong correspondence to the geographical relationships of specific countries or regions of origin. Clearer separation of different ethnic and regional populations was observed when northern and southern European groups were considered separately and the PCA results were influenced by the inclusion or exclusion of different self-identified population groups including Ashkenazi Jewish, Sardinian and Orcadian ethnic groups. SNP AIM sets were identified that could distinguish the regional and ethnic population groups. Moreover, the studies demonstrated that most allele frequency differences between different European groups could be effectively controlled in analyses using these AIM sets. The European substructure AIMs should be widely applicable to ongoing studies to confirm and delineate specific disease susceptibility candidate regions without the necessity to perform additional genome-wide SNP studies in additional subject sets.
Link
Human Y-chromosome mutation rate
- Even when we are able to routinely sequence entire Y chromosomes, we will still not generally be able to tell apart fathers and sons as sons (via their Y chromosome) only sometimes have different Y chromosomes than their fathers. In a large number of cases there will be no mutations.
- On the positive side, relationships in the entire human Y-chromosome phylogeny will be resolved with an accuracy of a few generations. We will no longer have to rely on poor resolution Y-STR "genetic distance" methods to determine relatedness of individuals. The chunky huge haplogroups dominating modern Y-chromosome pools will be resolved into a hierarchy of more exclusive families and really precise patterns of migration and admixture may be inferred.
Human Y Chromosome Base-Substitution Mutation Rate Measured by Direct Sequencing in a Deep-Rooting Pedigree
Yali Xue et al.
Abstract
Understanding the key process of human mutation is important for many aspects of medical genetics and human evolution. In the past, estimates of mutation rates have generally been inferred from phenotypic observations or comparisons of homologous sequences among closely related species [1,2,3]. Here, we apply new sequencing technology to measure directly one mutation rate, that of base substitutions on the human Y chromosome. The Y chromosomes of two individuals separated by 13 generations were flow sorted and sequenced by Illumina (Solexa) paired-end sequencing to an average depth of 11
 or 20
or 20 , respectively [4]. Candidate mutations were further examined by capillary sequencing in cell-line and blood DNA from the donors and additional family members. Twelve mutations were confirmed in
, respectively [4]. Candidate mutations were further examined by capillary sequencing in cell-line and blood DNA from the donors and additional family members. Twelve mutations were confirmed in  10.15 Mb; eight of these had occurred in
10.15 Mb; eight of these had occurred in vitro and four in
vitro and four in vivo. The latter could be placed in different positions on the pedigree and led to a mutation-rate measurement of 3.0
vivo. The latter could be placed in different positions on the pedigree and led to a mutation-rate measurement of 3.0
 10
10 8 mutations/nucleotide/generation (95% CI: 8.9
8 mutations/nucleotide/generation (95% CI: 8.9  10
10 9
9 7.0
7.0  10
10 8), consistent with estimates of 2.3
8), consistent with estimates of 2.3  10
10 8
8 6.3
6.3  10
10 8 mutations/nucleotide/generation for the same Y-chromosomal region from published human-chimpanzee comparisons [5] depending on the generation and split times assumed.
8 mutations/nucleotide/generation for the same Y-chromosomal region from published human-chimpanzee comparisons [5] depending on the generation and split times assumed.Link
August 26, 2009
Bronze Age origin of Semitic languages
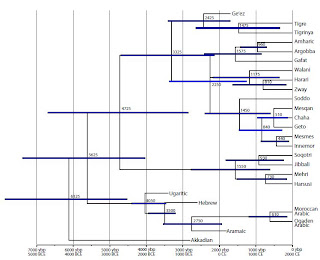 Bayesian phylogenetic methods, originally developed for biology, have been increasingly -and successfully- applied to linguistic data in recent years (e.g., for Indo-Europeans, Melanesians, and Austronesian speakers from the Pacific).
Bayesian phylogenetic methods, originally developed for biology, have been increasingly -and successfully- applied to linguistic data in recent years (e.g., for Indo-Europeans, Melanesians, and Austronesian speakers from the Pacific).The current paper proposes a Bronze Age origin for Semitic languages, ~3 thousand years after the split of European from Anatolian Indo-European speakers. I don't find this particularly surprising, as Semitic has been, until relatively recently, much more geographically constrained than Indo-European, and -due to the early literacy of the populations of the Near East, its post-Neolithic arrival can be observed in the archaeological record itself.
The current study also demonstrates that linguistic Bayesian phylogenetics (LBP) has no inherent bias to produce older dates for language dispersals; while the origin of the Indo-European (IE) language family has been dated to the early European Neolithic, and now Semitic to ~6,000 years, the spread of Melanesian languages to Pleistocene times, and of the Austronesian settlement of the Pacific to ~5,000 years. The congruence between LBP and traditional archaeology in all these cases should force IE exceptionalists who cling to the old theory of "steppe horse riders" to explain why, only in the dispersal of IE, it should LBP should have failed.
The paper also has free supplementary data, including a multistate phylogeny (pdf) of Semitic languages (reproduced top left of this post).
Furthermore, Eblaite (no Eblaite wordlists were available for our study), the closest relative of Akkadian and the only other member of East Semitic, was spoken in the Levant (specifically the northeast Levant or present-day Syria; Gordon 1997), which is also where some of the oldest West Semitic languages were spoken (Ugaritic, Aramaic and ancient Hebrew). The presence of ancient members of the two oldest Semitic groups (East andWest Semitic) in the same region of the Levant, combined with a possible long interval (100–3000 years) between the origin of Semitic and the appearance of Akkadian in Sumer, suggests a Semitic origin in the northeast Levant and a later movement of Akkadian eastward into Mesopotamia and Sumer (see figure 1 for a map of our proposed Semitic dispersals).
Lacking closely related non-Semitic languages to serve as out-groups in our phylogeny, we cannot estimate when or where the ancestor of all Semitic languages diverged from Afroasiatic. Furthermore, it is likely that some early Semitic languages became extinct and left no record of their existence. This is especially probable if early Semitic societies were pastoralist in nature (Blench 2006), as pastoralists are less likely to leave epigraphic and archaeological evidence of their languages.
Proc. R. Soc. B 7 August 2009 vol. 276 no. 1668 2703-2710
Bayesian phylogenetic analysis of Semitic languages identifies an Early Bronze Age origin of Semitic in the Near East
Andrew Kitchen et al.
Abstract
The evolution of languages provides a unique opportunity to study human population history. The origin of Semitic and the nature of dispersals by Semitic-speaking populations are of great importance to our understanding of the ancient history of the Middle East and Horn of Africa. Semitic populations are associated with the oldest written languages and urban civilizations in the region, which gave rise to some of the world's first major religious and literary traditions. In this study, we employ Bayesian computational phylogenetic techniques recently developed in evolutionary biology to analyse Semitic lexical data by modelling language evolution and explicitly testing alternative hypotheses of Semitic history. We implement a relaxed linguistic clock to date language divergences and use epigraphic evidence for the sampling dates of extinct Semitic languages to calibrate the rate of language evolution. Our statistical tests of alternative Semitic histories support an initial divergence of Akkadian from ancestral Semitic over competing hypotheses (e.g. an African origin of Semitic). We estimate an Early Bronze Age origin for Semitic approximately 5750 years ago in the Levant, and further propose that contemporary Ethiosemitic languages of Africa reflect a single introduction of early Ethiosemitic from southern Arabia approximately 2800 years ago.
Link
Demic diffusion of agriculture into Europe supported by craniometric data
 A previous article by Pinhasi et al. Tracing the Origin and Spread of Agriculture in Europe
A previous article by Pinhasi et al. Tracing the Origin and Spread of Agriculture in EuropePLoS ONE 4(8): e6747. doi:10.1371/journal.pone.0006747
Craniometric Data Supports Demic Diffusion Model for the Spread of Agriculture into Europe
Ron Pinhasi, Noreen von Cramon-Taubadel
Abstract
Background
The spread of agriculture into Europe and the ancestry of the first European farmers have been subjects of debate and controversy among geneticists, archaeologists, linguists and anthropologists. Debates have centred on the extent to which the transition was associated with the active migration of people as opposed to the diffusion of cultural practices. Recent studies have shown that patterns of human cranial shape variation can be employed as a reliable proxy for the neutral genetic relationships of human populations.
Methodology/Principal Findings
Here, we employ measurements of Mesolithic (hunter-gatherers) and Neolithic (farmers) crania from Southwest Asia and Europe to test several alternative population dispersal and hunter-farmer gene-flow models. We base our alternative hypothetical models on a null evolutionary model of isolation-by-geographic and temporal distance. Partial Mantel tests were used to assess the congruence between craniometric distance and each of the geographic model matrices, while controlling for temporal distance. Our results demonstrate that the craniometric data fit a model of continuous dispersal of people (and their genes) from Southwest Asia to Europe significantly better than a null model of cultural diffusion.
Conclusions/Significance
Therefore, this study does not support the assertion that farming in Europe solely involved the adoption of technologies and ideas from Southwest Asia by indigenous Mesolithic hunter-gatherers. Moreover, the results highlight the utility of craniometric data for assessing patterns of past population dispersal and gene flow.
Link
Lower Paleolithic hunters from Qesem Cave
"The Lower Paleolithic (earlier) hunters were skilled hunters of large game animals, as were Upper Paleolithic (later) humans at this site," UA anthropology professor Mary C. Stiner said.PNAS doi:10.1073/pnas.0900564106
"This might not seem like a big deal to the uninitiated, but there's a lot of speculation as to whether people of the late Lower Paleolithic were able to hunt at all, or whether they were reduced to just scavenging," Stiner said. "Evidence from Qesem Cave says that just like later Paleolithic humans, the earlier Paleolithic humans focused on harvesting large game. They were really at the top of the food chain."
Cooperative hunting and meat sharing 400–200 kya at Qesem Cave, Israel
Mary C. Stiner et al.
Abstract
Zooarchaeological research at Qesem Cave, Israel demonstrates that large-game hunting was a regular practice by the late Lower Paleolithic period. The 400- to 200,000-year-old fallow deer assemblages from this cave provide early examples of prime-age-focused ungulate hunting, a human predator–prey relationship that has persisted into recent times. The meat diet at Qesem centered on large game and was supplemented with tortoises. These hominins hunted cooperatively, and consumption of the highest quality parts of large prey was delayed until the food could be moved to the cave and processed with the aid of blade cutting tools and fire. Delayed consumption of high-quality body parts implies that the meat was shared with other members of the group. The types of cut marks on upper limb bones indicate simple flesh removal activities only. The Qesem cut marks are both more abundant and more randomly oriented than those observed in Middle and Upper Paleolithic cases in the Levant, suggesting that more (skilled and unskilled) individuals were directly involved in cutting meat from the bones at Qesem Cave. Among recent humans, butchering of large animals normally involves a chain of focused tasks performed by one or just a few persons, and butchering guides many of the formalities of meat distribution and sharing that follow. The results from Qesem Cave raise new hypotheses about possible differences in the mechanics of meat sharing between the late Lower Paleolithic and Middle Paleolithic.
Link
August 25, 2009
John Hawks on historical trends in length of life
Syllogistically speaking, Socrates didn't die of natural causes, therefore the Greeks had lifespans the same as ours. Or something.
...
But there's no doubt that Romans, Egyptians, and Greeks were dropping dead at age 30, 40, 50 and 60 -- at much higher age-specific mortality rates than today. Estimating the overall age profile is difficult and requires models.
Fortunately, there has been a study of the length of life in ancient Greece, that shows, that Socrates was not a unique case:
In a study of all men of renown, living in the 5th and 4th century in Greece, we identified 83 whose date of birth and death have been recorded with certainty. Their mean +/- SD and median lengths of life were found to be 71.3+/-13.4 and 70 years, respectively.
Of course, the lifespan of "men of renown" should be correlated with that of the general population, but with a higher mean, since men require time to achieve "renown". But, certainly the figure of 71 years does not seem too different from that of more recent men of renown, which is perhaps more surprising if one accounts for the high levels of violence in ancient Greece.
UPDATE I
John also writes:
More important, we don't have a clue what the maximum lifespan may have been 200, 500, or 2000 years ago. Such a tiny fraction of people make it above age 100 today that we could hardly expect to find any of them at all from skeletal samples. Nor can we expect accurate ages from historical records -- Methuseleh, anyone?
We do have some fairly secure dates for at least some ancient individuals. Strabo died at 88. Sophocles died at 90. Democritus' life span was said to be anything from 90-109. Alexis, the comic poet reached the age of 100, Isocrates, 98. So, there were probably centenarians in ancient times, although what the maximum was is anyone's guess.
UPDATE II
Psalm 90:10 gives the length of life as 70.
The days of our years are three score years and ten; and if by reason of strength they be fourscore years, yet is their strength labor and sorrow; for it is soon cut off, and we fly away.
Herodotus (1.32) has Solon give the "limit of life" at 70.
Croesus, you ask me about human affairs, and I know that the divine is entirely grudging and troublesome to us. [2] In a long span of time it is possible to see many things that you do not want to, and to suffer them, too. I set the limit of a man's life at seventy years;This suggests to me that a length of life of 70 was formulaic in the ancient Mediterranean, and corresponds quite well with the average recorded length for prominent Greeks.
The reproductive function in men usually continues active till they are sixty years old; if they pass beyond this period, till they are seventy; and some men have had children at seventy years old.
Women should marry when they are about eighteen years of age, and men at seven and thirty.The prescription to marry at 37, suggests that early death must not have been too common; otherwise, it would be difficult to maintain the population's numbers, a topic which clearly occupied Aristotle, who commented on Sparta's oligandria in the same work.
Of the orators, Gorgias, whom some call a sophist, lived to be one hundred and eight, and starved himself to death. They say that when he was asked the reason for his great age, sound in all his faculties, he replied that he had never accepted other peoples invitations to dinner!
Of the historians, Ctesibius died at the age of one hundred and four while taking a walk, according to Apollodorus in his Chronology.
Hieronymus, who went to war and stood much toil and many wounds, lived one hundred and four years, as Agatharchides says in the ninth book of his History of Asia; and he expresses his amazement at the man, because up to his last day he was still vigorous in his marital relations and in all his faculties, lacking none of the symptoms of health.
A reader writes in the comments:
The maximum life expectancy for humans seems to have been about 120years throughout recorded history.
"My Spirit will not strive with man forever, for he is indeed flesh; yet his days shall be one hundred and twenty years." Genesis 6:3
"From Cicero’s death to our day is a hundred and twenty years, one man’s life-time." Tacitus, "A Dialogue on Oratory", 17. Tacitus writes of seeing a 120 year old man in Britain in the 1st century A.D.
120 years as a length of exceptionally long-lived people is also mentioned by Herodotus:
The Icthyophagi then in their turn questioned the king concerning the term of life, and diet of his people, and were told that most of them lived to be a hundred and twenty years old, while some even went beyond that age- they ate boiled flesh, and had for their drink nothing but milk.
Of course we can't take the veracity of this story at face value, but the choice of number may suggest that it is as great as it could be without becoming unbelievable to the audience.
Admixture in northeastern Mexico
Ancestry informative markers and admixture proportions in northeastern Mexico
Martinez-Fierro ML, Beuten J, Leach RJ, Parra EJ, Cruz-Lopez M, Rangel-Villalobos H, Riego-Ruiz LR, Ortiz-Lopez R, Martinez-Rodriguez HG, Rojas-Martinez A.
To investigate the ancestral admixture in the Mestizo population in northeastern Mexico, we genotyped 74 ancestral informative markers (AIMs) and 15 Y-single-nucleotide polymorphisms (Y-SNPs) in 100 individuals. The Native American contribution is 56% (range: 27.4-81.2%), the European contribution is 38% (range: 16.7-70.5%) and the West African contribution is 6%. The results show a higher European contribution than was reported in other similar studies in the country, albeit with a predominant Native American ancestry. No remarkable differences in the ancestry proportions were observed using subgroups of 74, 54, 34 and 24 AIMs. The paternal lineage calculated by genotyping of 15 Y-SNPs, shows a major component of European and Eurasian ancestry markers ( approximately 78%), compared with Amerindian ( approximately 12%) and African markers (10%). This information will set a reference for future determinations of admixture proportions in the Mestizo population from Mexico and for population-based association studies of complex diseases.
Link
August 24, 2009
Amerindian mtDNA in Argentinean population
Amerindian mitochondrial DNA haplogroups predominate in the population of Argentina: towards a first nationwide forensic mitochondrial DNA sequence database.
Bobillo MC, Zimmermann B, Sala A, Huber G, Röck A, Bandelt HJ, Corach D, Parson W.
The study presents South American mitochondrial DNA (mtDNA) data from selected north (N = 98), central (N = 193) and south (N = 47) Argentinean populations. Sequence analysis of the complete mtDNA control region (CR, 16024-576) resulted in 288 unique haplotypes ignoring C-insertions around positions 16193, 309, and 573; the additional analysis of coding region single nucleotide polymorphisms enabled a fine classification of the described lineages. The Amerindian haplogroups were most frequent in the north and south representing more than 60% of the sequences. A slightly different situation was observed in central Argentina where the Amerindian haplogroups represented less than 50%, and the European contribution was more relevant. Particular clades of the Amerindian subhaplogroups turned out to be nearly region-specific. A minor contribution of African lineages was observed throughout the country. This comprehensive admixture of worldwide mtDNA lineages and the regional specificity of certain clades in the Argentinean population underscore the necessity of carefully selecting regional samples in order to develop a nationwide mtDNA database for forensic and anthropological purposes. The mtDNA sequencing and analysis were performed under EMPOP guidelines in order to attain high quality for the mtDNA database.
Link
August 23, 2009
Comparison of Neanerthal and modern human diets
Isotopic evidence for the diets of European Neanderthals and early modern humans
Michael P. Richards, Erik Trinkaus
Abstract
We report here on the direct isotopic evidence for Neanderthal and early modern human diets in Europe. Isotopic methods indicate the sources of dietary protein over many years of life, and show that Neanderthals had a similar diet through time (≈120,000 to ≈37,000 cal BP) and in different regions of Europe. The isotopic evidence indicates that in all cases Neanderthals were top-level carnivores and obtained all, or most, of their dietary protein from large herbivores. In contrast, early modern humans (≈40,000 to ≈27,000 cal BP) exhibited a wider range of isotopic values, and a number of individuals had evidence for the consumption of aquatic (marine and freshwater) resources. This pattern includes Oase 1, the oldest directly dated modern human in Europe (≈40,000 cal BP) with the highest nitrogen isotope value of all of the humans studied, likely because of freshwater fish consumption. As Oase 1 was close in time to the last Neanderthals, these data may indicate a significant dietary shift associated with the changing population dynamics of modern human emergence in Europe.
Link
August 22, 2009
Carleton Coon on video
Here is the first episode:
Diet in southern French Neolithic populations
Southern French Neolithic populations: Isotopic evidence for regional specificities in environment and diet
Estelle Herrscher, Gwenaëlle Le Bras-Goude
Abstract
 4500-3500 BC cal) is characterized by the development of food production techniques as well as by increasing social complexity. These characteristics could have had an impact on human dietary patterns. To evaluate human dietary practices and lifeways of the Middle Neolithic populations from the South of France, stable carbon and nitrogen isotope analysis was carried out on 57 human and 53 faunal bones from seven archaeological sites located in the Languedoc and Garonne regions between 20 and 100 km from the Mediterranean Sea, respectively. Results show regional differences in carbon isotope values. Animal and human bones from the Languedoc region are significantly enriched in 13C relative to the Garonne. Conversely, human and dog bones from the Garonne region are significantly enriched in 15N compared to human and dog bones from the Languedoc region. These results highlight the importance of the local ecosystem in human and animal diet as well as a regional differentiation of palaeodietary behavior, which probably relates to economic and social factors. The comparison of stable isotope data with archaeological and biological evidence does not show any significant intra- or interpopulation differences. However, the presence of human outliers suggests that migration probably occurred, perhaps in relation to the trade of animals and/or materials. This study also highlights the importance of investigating local animal stable isotope values for the interpretation of human palaeodiet.
4500-3500 BC cal) is characterized by the development of food production techniques as well as by increasing social complexity. These characteristics could have had an impact on human dietary patterns. To evaluate human dietary practices and lifeways of the Middle Neolithic populations from the South of France, stable carbon and nitrogen isotope analysis was carried out on 57 human and 53 faunal bones from seven archaeological sites located in the Languedoc and Garonne regions between 20 and 100 km from the Mediterranean Sea, respectively. Results show regional differences in carbon isotope values. Animal and human bones from the Languedoc region are significantly enriched in 13C relative to the Garonne. Conversely, human and dog bones from the Garonne region are significantly enriched in 15N compared to human and dog bones from the Languedoc region. These results highlight the importance of the local ecosystem in human and animal diet as well as a regional differentiation of palaeodietary behavior, which probably relates to economic and social factors. The comparison of stable isotope data with archaeological and biological evidence does not show any significant intra- or interpopulation differences. However, the presence of human outliers suggests that migration probably occurred, perhaps in relation to the trade of animals and/or materials. This study also highlights the importance of investigating local animal stable isotope values for the interpretation of human palaeodiet.August 21, 2009
Post-glacial recolonization of Britain happened after 14,700 years BP
Volume 28, Issues 19-20, September 2009, Pages 1895-1913
doi:10.1016/j.quascirev.2009.03.006
The early Lateglacial re-colonization of Britain: new radiocarbon evidence from Gough's Cave, southwest England
R.M. Jacobi and T.F.G. Higham
Abstract
Gough's Cave is still Britain's most significant Later Upper Palaeolithic site. New ultrafiltered radiocarbon determinations on bone change our understanding of its occupation, by demonstrating that this lasted for only a very short span of time, at the beginning of the Lateglacial Interstadial (Greenland Interstadial 1 (GI-1: Bølling and Allerød)). The application of Bayesian modelling to the radiocarbon dates from this, and other sites from the period in southwest England, suggests that re-colonization after the Last Glacial Maximum took place only after 14,700 cal BP, and is, therefore, more recent than that of the Paris Basin and the Belgian Ardennes. On their own, the radiocarbon determinations cannot tell us whether re-colonization was synchronous with, just prior to, or after, Lateglacial warming. Isotopic studies of humanly-modified mammalian tooth enamel may be one way forward.
Link
August 20, 2009
John Hawks on Anne Wojcicki on Race
A lot of the difficulty in talking about race has been a lack of agreement on what “race” means. In the past, the idea of pure races also included an ordering of certain races as inherently superior to others. We reject this idea absolutely. However, that doesn’t mean that there are no genetic differences between populations of different ancestral origin. A few of our features use the genome-wide data of reference populations from around the world to trace the origin of pieces of an individual’s genome. Some customers have complex patterns depending on where their ancestors originated. These reference populations aren’t “races”; they’re representative samples of peoples who have lived in a single place for a very long time and have thus accumulated different sets of genetic variants over time.
That's a tricky piece of wordcraft -- they're not 'races'; "they're representative samples of peoples who have lived in a single place for a very long time and have thus accumulated different sets of genetic variants over time."
Uhh....I'm thinking that's pretty much the definition of race in a lot of textbooks...
The points she starts with -- both true -- are that human populations aren't isolated ("pure races"), and you shouldn't rank them ("inherently superior"). But those ideas conflict with the process, since the software commonly used in human genetics (like STRUCTURE and other programs) assumes a model in which originally isolated groups (otherwise known as pure races) mix together.
It is true that STRUCTURE (and similar tools) use a model in which each individual is assumed to have a share of genes from each of K different populations, that differ from each other in the frequency (and in some case co-occurrence, due to linkage) of different gene variants.
These K populations, are not, however, assumed to be "pure races", but rather simply populations that differ from each other in their genetic characteristics. The program itself does not make any assumptions as to why they differ from each other: it could be due to different types or intensity of gene flow, or to reproductive isolation. Distinctive gene frequencies may arise both due to "race purity" or in the presence of gene flow, provided that it occurs at a low enough level so that changes that occur in one population are not reflected immediately in the other, and their distinctiveness is maintained.
But, more importantly, the "pure races" model is an approximation whose validity can be checked. If gene flow is substantial, then our simplifying STRUCTURE view of the world will not result in distinct clusters, in which the great majority of individuals from a particular population have the greatest share of their ancestry from the same racial clusters.
In other words, the fact that we assume distinct "pure" clusters is no guarantee that we will get distinct races via an application of STRUCTURE. Try STRUCTURE with K=2 in a homogeneous population, and you won't get 2 distinct clusters, even though you made that assumption: you will get a bunch of individuals that belong -in various proportions- to the 2 clusters, but no 2 solid blocks of individuals, one of which belongs predominantly to the first, and the other to the second cluster.
What STRUCTURE and similar programs have repeatedly shown, is that humans do, in fact, have genomes that resemble each other in the way we would expect if there were pure races which have occasionally mixed in their peripheries.
Our understanding of genetic processes helps us understand, that this is not the result of separate creation of human races, but of long-term gene flow limitations due to geography and culture, that have allowed originally related human populations to evolve apart.
Male-female differences in craniofacial dimensions of Koreans
Female-to-male proportions of the head and face in Koreans.
Song WC, Kim JI, Kim SH, Shin DH, Hu KS, Kim HJ, Lee JY, Koh KS.
It is well known that the head and face are smaller in female subjects than in male subjects. However, almost all previous studies have quantified the size difference between female and male subjects as simple numerical values, which might not clarify the difference. The present study evaluated the female-to-male proportions of the head and face so as to clarify the sex-related differences. A total of 1939 female subjects and 1398 male subjects were divided into 3 age groups: young (20-39 y), middle-aged (40-59 y), and elderly (60-79 y). The dimensions were classified into 3 categories: 5 cephalic, 3 frontal facial, and 6 lateral facial. The female-to-male proportions of individual dimensions were compared in the 3 age groups using the following formula: female measurement value x 100/(mean of male measurement value). The female-to-male proportions of the cephalic dimension increased with age, with the female cephalic dimensions overall being about 96% of the male cephalic dimensions. The female-to-male proportions of the frontal facial dimension were constant across the age groups, with the female frontal facial dimensions overall being 95% of the male frontal facial dimensions. The female lateral facial dimension increased markedly from the young to middle-aged group and was constant or decreased slightly from the middle-aged to the elderly group. Overall, the female lateral facial dimensions were approximately 97% of the male lateral facial dimensions. The present study will suggest a new approach to elucidate those sex-related dimensional differences that are characteristic of female and male subjects.
Link
August 19, 2009
East vs. West differences in facial expressions
Current Biology, doi:10.1016/j.cub.2009.07.051
Cultural Confusions Show that Facial Expressions Are Not Universal
Rachael E. Jack
Abstract
Central to all human interaction is the mutual understanding of emotions, achieved primarily by a set of biologically rooted social signals evolved for this purposefacial expressions of emotion. Although facial expressions are widely considered to be the universal language of emotion [1,2,3], some negative facial expressions consistently elicit lower recognition levels among Eastern compared to Western groups (see [4] for a meta-analysis and [5,6] for review). Here, focusing on the decoding of facial expression signals, we merge behavioral and computational analyses with novel spatiotemporal analyses of eye movements, showing that Eastern observers use a culture-specific decoding strategy that is inadequate to reliably distinguish universal facial expressions of fear and disgust. Rather than distributing their fixations evenly across the face as Westerners do, Eastern observers persistently fixate the eye region. Using a model information sampler, we demonstrate that by persistently fixating the eyes, Eastern observers sample ambiguous information, thus causing significant confusion. Our results question the universality of human facial expressions of emotion, highlighting their true complexity, with critical consequences for cross-cultural communication and globalization.
Link
August 18, 2009
Mediterranean diet, physical activity, and Alzheimer's disease
JAMA 302(6):627-637.
Physical Activity, Diet, and Risk of Alzheimer Disease
Nikolaos Scarmeas et al.
Abstract<
Context Both higher adherence to a Mediterranean-type diet and more physical activity have been independently associated with lower Alzheimer disease (AD) risk but their combined association has not been investigated.
Objective To investigate the combined association of diet and physical activity with AD risk.
Design, Setting, and Patients Prospective cohort study of 2 cohorts comprising 1880 community-dwelling elders without dementia living in New York, New York, with both diet and physical activity information available. Standardized neurological and neuropsychological measures were administered approximately every 1.5 years from 1992 through 2006. Adherence to a Mediterranean-type diet (scale of 0-9; trichotomized into low, middle, or high; and dichotomized into low or high) and physical activity (sum of weekly participation in various physical activities, weighted by the type of physical activity [light, moderate, vigorous]; trichotomized into no physical activity, some, or much; and dichotomized into low or high), separately and combined, were the main predictors in Cox models. Models were adjusted for cohort, age, sex, ethnicity, education, apolipoprotein E genotype, caloric intake, body mass index, smoking status, depression, leisure activities, a comorbidity index, and baseline Clinical Dementia Rating score.
Main Outcome Measure Time to incident AD.
Results A total of 282 incident AD cases occurred during a mean (SD) of 5.4 (3.3) years of follow-up. When considered simultaneously, both Mediterranean-type diet adherence (compared with low diet score, hazard ratio [HR] for middle diet score was 0.98 [95% confidence interval {CI}, 0.72-1.33]; the HR for high diet score was 0.60 [95% CI, 0.42-0.87]; P = .008 for trend) and physical activity (compared with no physical activity, the HR for some physical activity was 0.75 [95% CI, 0.54-1.04]; the HR for much physical activity was 0.67 [95% CI, 0.47-0.95]; P = .03 for trend) were associated with lower AD risk. Compared with individuals neither adhering to the diet nor participating in physical activity (low diet score and no physical activity; absolute AD risk of 19%), those both adhering to the diet and participating in physical activity (high diet score and high physical activity) had a lower risk of AD (absolute risk, 12%; HR, 0.65 [95% CI, 0.44-0.96]; P = .03 for trend).
Conclusion In this study, both higher Mediterranean-type diet adherence and higher physical activity were independently associated with reduced risk for AD.
Link
August 17, 2009
Coastal-inland differences in Y chromosomes of the Levant

Inspit of these deficiencies, we may still make some useful observations. For example, IE-speaking Iranians have largely the same haplogroups as Arabs, but a much higher representation of haplogroup J2 compared to J1. The converse is true for all Arabs except the Lebanese. But, we do know, that even in Lebanon itself, Muslims have a higher J1/J2 ratio than Christians, and Islam was the main vehicle of Arabization in the region. The Christians are descended from the pre-Arab Byzantine Greco-Aramaic populations (with an addition of Western European Y-chromosomes in some Christian communities, which would not have substantially upset the J1/J2 balance).
Annals of Human Genetics doi:10.1111/j.1469-1809.2009.00538.x
Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast
Mirvat El-Sibai et al.
Abstract
We have examined the male-specific phylogeography of the Levant and its surroundings by analyzing Y-chromosomal haplogroup distributions using 5874 samples (885 new) from 23 countries. The diversity within some of these haplogroups was also examined. The Levantine populations showed clustering in SNP and STR analyses when considered against a broad Middle-East and North African background. However, we also found a coastal-inland, east-west pattern of diversity and frequency distribution in several haplogroups within the small region of the Levant. Since estimates of effective population size are similar in the two regions, this strong pattern is likely to have arisen mainly from differential migrations, with different lineages introduced from the east and west.
Link
Malaria and GYPC selection in humans vs. other primates
Molecular evolution of GYPC: Evidence for recent structural innovation and positive selection in humans
Jason A. Wilder et al.
Abstract
GYPC encodes two erythrocyte surface sialoglycoproteins in humans, glycophorin C and glycophorin D (GPC & GPD), via initiation of translation at two start codons on a single transcript. The malaria-causing parasite Plasmodium falciparum uses GPC as a means of invasion into the human red blood cell. Here we examine the molecular evolution of GYPC among the Hominoidea (Greater and Lesser Apes) and also the pattern of polymorphism at the locus in a global human sample. We find an excess of non-synonymous divergence among species that appears to be caused solely by accelerated evolution of GYPC in the human lineage. Moreover, we find that the ability of GYPC to encode both GPC and GPD is a uniquely human trait, caused by the evolution of the GPC start codon in the human lineage. The pattern of polymorphism among humans is consistent with a hitchhiking event at the locus, suggesting that positive natural selection affected GYPC in the relatively recent past. Because GPC is exploited by P. falciparum for invasion of the red blood cell, we hypothesize that selection for evasion of P. falciparum has caused accelerated evolution of GYPC in humans (relative to other primates), and that this positive selection has continued to act in the recent evolution of our species. These data suggest that malaria has played a powerful role in shaping molecules on the surface of the human red blood cell. In addition, our examination of GYPC reveals a novel mechanism of protein evolution: co-option of UTR sequence following the formation of a new start codon. In the case of human GYPC the ancestral protein (GPD) continues to be produced through leaky translation. Because leaky translation is a widespread phenomenon among genes and organisms, we suggest that co-option of UTR sequence may be an important source of protein innovation.
Link
August 16, 2009
72 thousand year old heat-treated tools from South Africa
Science doi:10.1126/science.1175028
Fire As an Engineering Tool of Early Modern Humans
Kyle S. Brown et al.
Abstract
The controlled use of fire was a breakthrough adaptation in human evolution. It first provided heat and light and later allowed the physical properties of materials to be manipulated for the production of ceramics and metals. The analysis of tools at multiple sites shows that the source stone materials were systematically manipulated with fire to improve their flaking properties. Heat treatment predominates among silcrete tools at ~72 thousand years ago (ka) and appears as early as 164 ka at Pinnacle Point, on the south coast of South Africa. Heat treatment demands a sophisticated knowledge of fire and an elevated cognitive ability and appears at roughly the same time as widespread evidence for symbolic behavior.
Link
August 14, 2009
Genome-wide STRs and American prehistory
Hierarchical modeling of genome-wide Short Tandem Repeat (STR) markers infers native American prehistory
Cecil M. Lewis Jr.
Abstract
This study examines a genome-wide dataset of 678 Short Tandem Repeat loci characterized in 444 individuals representing 29 Native American populations as well as the Tundra Netsi and Yakut populations from Siberia. Using these data, the study tests four current hypotheses regarding the hierarchical distribution of neutral genetic variation in native South American populations: (1) the western region of South America harbors more variation than the eastern region of South America, (2) Central American and western South American populations cluster exclusively, (3) populations speaking the Chibchan-Paezan and Equatorial-Tucanoan language stock emerge as a group within an otherwise South American clade, (4) Chibchan-Paezan populations in Central America emerge together at the tips of the Chibchan-Paezan cluster. This study finds that hierarchical models with the best fit place Central American populations, and populations speaking the Chibchan-Paezan language stock, at a basal position or separated from the South American group, which is more consistent with a serial founder effect into South America than that previously described. Western (Andean) South America is found to harbor similar levels of variation as eastern (Equatorial-Tucanoan and Ge-Pano-Carib) South America, which is inconsistent with an initial west coast migration into South America. Moreover, in all relevant models, the estimates of genetic diversity within geographic regions suggest a major bottleneck or founder effect occurring within the North American subcontinent, before the peopling of Central and South America.
Link
August 13, 2009
Artificial selection in dairy cattle
The Genome Response to Artificial Selection: A Case Study in Dairy Cattle
Laurence Flori et al.
Abstract
Dairy cattle breeds have been subjected over the last fifty years to intense artificial selection towards improvement of milk production traits. In this study, we performed a whole genome scan for differentiation using 42,486 SNPs in the three major French dairy cattle breeds (Holstein, Normande and Montbéliarde) to identify the main physiological pathways and regions which were affected by this selection. After analyzing the population structure, we estimated FST within and across the three breeds for each SNP under a pure drift model. We further considered two different strategies to evaluate the effect of selection at the genome level. First, smoothing FST values over each chromosome with a local variable bandwidth kernel estimator allowed identifying 13 highly significant regions subjected to strong and/or recent positive selection. Some of them contained genes within which causal variants with strong effect on milk production traits (GHR) or coloration (MC1R) have already been reported. To go further in the interpretation of the observed signatures of selection we subsequently concentrated on the annotation of differentiated genes defined according to the FST value of SNPs localized close or within them. To that end we performed a comprehensive network analysis which suggested a central role of somatotropic and gonadotropic axes in the response to selection. Altogether, these observations shed light on the antagonism, at the genome level, between milk production and reproduction traits in highly producing dairy cows.
Link
August 12, 2009
Ethnicity inference from DNA in Madrid terrorist attacks
PLoS ONE 4(8): e6583. doi:10.1371/journal.pone.0006583
Ancestry Analysis in the 11-M Madrid Bomb Attack Investigation
Christopher Phillips et al.
Abstract
The 11-M Madrid commuter train bombings of 2004 constituted the second biggest terrorist attack to occur in Europe after Lockerbie, while the subsequent investigation became the most complex and wide-ranging forensic case in Spain. Standard short tandem repeat (STR) profiling of 600 exhibits left certain key incriminatory samples unmatched to any of the apprehended suspects. A judicial order to perform analyses of unmatched samples to differentiate European and North African ancestry became a critical part of the investigation and was instigated to help refine the search for further suspects. Although mitochondrial DNA (mtDNA) and Y-chromosome markers routinely demonstrate informative geographic differentiation, the populations compared in this analysis were known to show a proportion of shared mtDNA and Y haplotypes as a result of recent gene-flow across the western Mediterranean, while any two loci can be unrepresentative of the ancestry of an individual as a whole. We based our principal analysis on a validated 34plex autosomal ancestry-informative-marker single nucleotide polymorphism (AIM-SNP) assay to make an assignment of ancestry for DNA from seven unmatched case samples including a handprint from a bag containing undetonated explosives together with personal items recovered from various locations in Madrid associated with the suspects. To assess marker informativeness before genotyping, we predicted the probable classification success for the 34plex assay with standard error estimators for a naïve Bayesian classifier using Moroccan and Spanish training sets (each n = 48). Once misclassification error was found to be sufficiently low, genotyping yielded seven near-complete profiles (33 of 34 AIM-SNPs) that in four cases gave probabilities providing a clear assignment of ancestry. One of the suspects predicted to be North African by AIM-SNP analysis of DNA from a toothbrush was identified late in the investigation as Algerian in origin. The results achieved illustrate the benefit of adding specialized marker sets to provide enhanced scope and power to an already highly effective system of DNA analysis for forensic identification.
Link
August 11, 2009
Finally, an updated look at Y-chromosomes of Jewish priests (Hammer et al. 2009)
Thus, it became necessary to find a more stringent characterization of Cohen Y-chromosomes that would represent true founder effects in that population. The new paper seems to identify at least two such lineages, one in J-P58, which is a subset of J1, and one in J2a-M410*, and estimates that they were founded 3.2 and 4.2 thousand years ago.
I will comment on this further when I get a hold of the paper and supplementary material.
UPDATE: The authors use the evolutionary mutation rate, and thus the presented ages are overestimated significantly. However, there are reasons to doubt the germline-rate estimate of about 1,000 years for the J1 lineage:
- Demographic plausibility of growth to encompass nearly a third the Cohanim in about 1,000 years. I am not sure what the demic size of Cohanim is, but going from 1 individual to the current population size would require a consistent high growth over many generations. This seems implausible, unless there is indeed historical evidence for such a Cohen founder's descendants extraordinary success.
- The presence of the founding lineage in both Ashkenazim and Sephardim may suggest a common ancestor before the separation of these two populations.
Pinpointing Jewish priestly founders to specific individuals, including Biblical ones, is not easy, as the Y-STR technology does not allow for anything resembling accurate age estimation. However, the paper is a welcome new study of a much-discussed topic, and adds significantly to our understanding.
UPDATE II: On the other hand, the J-P58* haplogroup was found in 325 of 2,099 non-Jews surveyed, but the extended Cohen Modal Haplotype (eCMH) in none (Table S2). If the eCMH founder lived 3+ kya, it would be strange indeed if he left no non-Jewish descendants, as it would imply zero conversion from that lineage to other religions. The lack of non-Jewish eCMHs does support the "Jewishness" of this lineage, but on the other hand, makes a very old age more difficult to accept.
My guess is that the eCMH founder lived in Roman times. This would simultaneously allow enough time to explain the lineage's geographical and demographic growth, while also explaining its limited penetration to non-Jewish populations.
Human Genetics doi:10.1007/s00439-009-0727-5
Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood
Michael F. Hammer et al.
Abstract
It has been known for over a decade that a majority of men who self report as members of the Jewish priesthood (Cohanim) carry a characteristic Y chromosome haplotype termed the Cohen Modal Haplotype (CMH). The CMH has since been used to trace putative Jewish ancestral origins of various populations. However, the limited number of binary and STR Y chromosome markers used previously did not provide the phylogenetic resolution needed to infer the number of independent paternal lineages that are encompassed within the Cohanim or their coalescence times. Accordingly, we have genotyped 75 binary markers and 12 Y-STRs in a sample of 215 Cohanim from diverse Jewish communities, 1,575 Jewish men from across the range of the Jewish Diaspora, and 2,099 non-Jewish men from the Near East, Europe, Central Asia, and India. While Cohanim from diverse backgrounds carry a total of 21 Y chromosome haplogroups, 5 haplogroups account for 79.5% of Cohanim Y chromosomes. The most frequent Cohanim lineage (46.1%) is marked by the recently reported P58 T->C mutation, which is prevalent in the Near East. Based on genotypes at 12 Y-STRs, we identify an extended CMH on the J-P58* background that predominates in both Ashkenazi and non-Ashkenazi Cohanim and is remarkably absent in non-Jews. The estimated divergence time of this lineage based on 17 STRs is 3,190 ± 1,090 years. Notably, the second most frequent Cohanim lineage (J-M410*, 14.4%) contains an extended modal haplotype that is also limited to Ashkenazi and non-Ashkenazi Cohanim and is estimated to be 4.2 ± 1.3 ky old. These results support the hypothesis of a common origin of the CMH in the Near East well before the dispersion of the Jewish people into separate communities, and indicate that the majority of contemporary Jewish priests descend from a limited number of paternal lineages.
Electronic supplementary material The online version of this article (doi:10.1007/s00439-009-0727-5) contains supplementary material, which is available to authorized users.
Link

